Abstract
Introduction: Most Philadelphia-positive (Ph+) Acute Lymphoblastic Leukemia (ALL) alleles are BCR1ABL1 p190 fusion transcripts. Objectives: Enhance and test acute lymphoblastic leukaemia BCR-ABL1 p190 transcript finding and tracking RT qPCR tool. Methodology: Cycle frequency, primer concentration, and real-time qPCR output, volume, and results improve acute lymphoblastic leukaemia data. Normal curve and element analysis needed enough cDNA. Before cDNA, Nanodrop tested distal blood RNA. Nanodrop RNA measures and extracts RNA. We used specialised gear and BCR-ABL P190 primers to find a genomic variety. Numerous samples were examined for accuracy. Trials lasted 1–3 days. Results: 15 BCR-ABL+ALL tissues had CT values around 25 cycles. The water stopped errors. Retested parts. 45 chronic myeloid and 15 acute lymphoblastic leukaemia samples were BCR-ABLp190-negative using the same method. Conclusion: Real-time PCR found BCR-ABL1. Better BCR-ABL1 p190 detection and lab accuracy allow real-time qPCR to track ALL.
Key Words
RT-qPCR, BCR ABL1 P190, Acute Lymphoblastic Leukemia, Detection
Introduction
A form of malignancy called leukaemia develops in the bone marrow and results in aberrant WBC. Blast or leukaemia cells are the terms used to describe these immature white blood cells. In 2020, it will account for 3.4% of all new instances of cancer and 3.8% of all cancer fatalities globally, ranking as the eleventh cause of carcinoma-related fatalities (Huang 2021).
Leukemia's fatality rate has decreased thanks to recently developed treatments like immunotherapy, point mutations focused antagonists, chimeric antigen receptor (CAR) T-cell therapy and apoptosis-inducing agents, yet it remains a common illness that results in disability and higher medical expenses (Martinez 2022).
Acute Leukemia is characterized by the malfunction of premature cells to differentiate into mature cells and the uncontrolled growth leading to its clinical manifestations. In the United States, the American Cancer Society’s estimates for ALL in 2022, the incidence of new cases was 6,660, comprises of males 3,740 and 2,920 females while the mortality rate is 1,560 deaths, comprising males (880 and 680 in females) (Saha 2023). In Pakistan's international agency for cancer, research leukaemia is in 5th position based on incidence with a total number of cases of 8305 (4.9%) and the mortality rate is 5th position. A study in KPK found district DIR having an incidence of 31 cases followed by Bajawar with 28 cases of ALL.
ALL can develop at any age, but the highest incidence is in children between the age range of 0-14 years. While in the mid of 20s, it declines and rises again after age 50. It increases after 50 years due to poor cytogenetics and comorbidities. In adults, it’s about 4/10 cases. In ALL the cancerous cell is lymphoblast. Regular lymphoblasts develop from disease (B-cells or T-cells), also known as lymphocytes, adult cells. Lymphocyte generation is managed by the organism. Both the regulation of the number of lymphoid cells and the proper development of lymphocytes are compromised in ALL.
PCR advantages include quickness and high sensitivity, Europe against Cancer (EAC) program conducted Initial efforts to standardize qRT-PCR analysis, the most frequent leukaemia-associated fusion genes were designed and validated taqpath primer and probe sets.
A study in Russia (2018) claimed that the detection of a BCR-ABL1-like profile by RT qPCR can be used as a fast and easy screening method. The p190 is used for the diagnosis and prognosis Of ALL (3, 4). The aim of my research is to develop the RT PCR assay for detecting the BCR-ABL1 P190 transcript in ALL patients so that clinicians can start effective therapy, such as tyrosine kinase inhibitors, as soon as possible to achieve remission and save the patient from more intensive chemotherapy. In addition, once refined, the RT PCR assay would be useful for tracking the P190 transcript in ALL patients who are currently receiving treatment. This will help the clinician assess the patient's current treatment and determine whether to continue, stop or change the treatment. The study's objective is to improve and confirm the RT qPCR method for the identification and tracking of BCR-ABL1 p190 transcript in individuals with ALL.
Methodology
An observation study was conducted at the Department of Hematology, Hayatabad medical complex. Non-probability convenient sampling was adopted and subjects were selected randomly from outdoor patients. From each patient, a five ml, peripheral blood (PB) sample was collected in tubes containing EDTA anticoagulant for detection of P190 BCR-ABL1. Out of a total 50 of samples, 15 samples were Philadelphia Positive Acute lymphoblastic leukaemia, 15 samples were Philadelphia Negative ALL and 30 samples were CML patients. Addition of Trizol. To Preserve RNA, after collecting blood, five ml of triazole reagent was added to preserve RNA and it is stored at (-80 degree Celsius). RNA Extraction: The sample was thawed on ice packs and vortexed. From this 1 ml of the sample was put into another Eppendorf tube followed by 7 ml of trizol reagent. It was thoroughly mixed. To allow the full breakdown of proteins of the nucleus complexes, the sample was warmed for five min at ambient temperature. To get rid of cell waste, it was subsequently spun.
It was put into the first Eppendorf tube. Separate the phases. For every 1 millilitre of TRIZOL Reagent, 250 ml of chloroform was introduced. It was Vortexed violently for fifteen sec and stored at room temperature for two to three minutes at a time. Materials were spun for fifteen min at 4 C using a maximum force of 13,000 x g. The combination was divided into darker crimson, a phenol-chloroform phase, interphase, and a translucent aqueous phase after spinning.
RNA Precipitation After the centrifuge, a clear liquid at the top is transferred to the second Eppendorf tube. To the second Eppendorf tube, add 100ul of chloroform was added and centrifuged in a refrigerated centrifuge at 13000xg for 14-15 min at 4 o'clock. Once more, the 3rd Eppendorf tube received the top watery layer. Propanol was mixed with the water phase to extract the RNA. For every 1 millilitre of the TRIZOL Reagent used for the initial grinding, use 700 ul of 2-propanol. The specimens were spun at a maximum of 12,000 x g for 10 minutes at 4 oC after being warmed for ten min at fifteen to thirty oC. RNA Wash: All of the liquid was eliminated. 700ul of 70 per cent ethanol was used to cleanse the RNA particle once more. Eppendorf was kept in a thermal block at 50 to 60 degrees Celsius for five minutes. RNA was incorporated in DEPC-treated water and combined with a few pipette tips' worths of solution. The concentration of 535.86 ng/ul extracted RNA was checked by nano-drop IBMS/PG/20 to know the quantity and quality of DNA. Thermofiscer cDNA kit: For cDNA preparation, a THERMOFISCER cDNA kit was used, the RNA sample was thawed in an ice bucket and vortexed, and the Reagents of the cDNA kit were thawed in an ice bucket. The detection and measurement of the expression of genes are done using RT-PCR (Quant Studio Version 5).
Results
The methodology section provides additional information regarding this study's unique real-time PCR strategy to identify CML and ALL. In order to successfully implement this method, many samples were collected. There were a total of sixty samples taken.
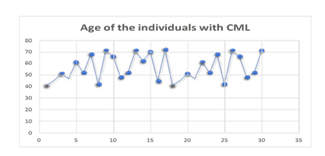
Thirty individuals out of the recruited samples were found to have Chronic Myeloid Leukemia. In addition, each and every diagnosed instance had tested positive for the Philadelphia chromosome. In addition, every person diagnosed with CML in this study was older than 40. The history of CML patients almost always included symptoms such as lethargy, fever, night sweats, and occasionally pallor, as illustrated in table 2. A comprehensive blood examination revealed an elevated number of white blood cells. In this study, the increased WBC count ranged anywhere from 150 x 109 to 280 x 10. In addition, patients diagnosed with CML did not have a history of specific medication use, nor do they have a history of receiving specific blood transfusions. The analysis of bone marrow revealed an increase in cellularity along with hyperplastic myeloid lineage.
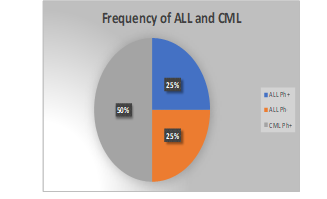
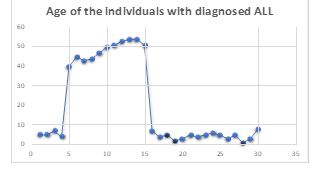
Within the scope of the present investigation, 30 cases have been identified of ALL. 15 of these 30 ALL instances were positive for the Philadelphia chromosome as confirmed by cytogenetics or already by PC, while the other 15 ALL cases tested negative for the Philadelphia chromosome. Figure 2 illustrates that out of the 15 cases of ALL with Philadelphia chromosomes, only two people were younger than ten years old. At the same time, the remaining thirteen participants are older than forty years old. Patients with a diagnosis of ALL who did not.
Philadelphia chromosomes aged younger than ten years are depicted in figure 2. Clinical manifestations of ALL, such as fever, bodily pains, and pallor, were present in almost every single affected individual. In addition, the ALL reduced the erythropoiesis, which led to a lower amount of HB, which ultimately resulted in the need for blood transfusion. Therefore, All these patients whether Philadelphia positive or negative, had a history of receiving a blood transfusion. As shown in Table 1, most persons with ALL screened positive for hepatomegaly, splenomegaly, and lymphadenopathy. As part of the diagnostic process, bone aspirations were performed to verify the existence of ALL the bone marrow smear revealed hypercellularity with blasts together with reduced suppressed megakaryopoiesis, erythropoiesis and myelopoiesis in addition, blasts stained negative for POX.
Philadelphia-positive acute lymphoblastic leukaemia (ALL) is a form of leukaemia that can affect anyone at any age if they have a copy of the Philadelphia chromosome. This discovery was made possible by recent research. It was found that the condition manifested itself in youngsters of a wide range of ages, beginning with those younger than 10 years old (2 cases). Not only was it discovered in younger people, but it was also discovered most frequently in people at least forty years old. Lymphadenopathy splenomegaly and hepatomegaly were found in the majority of these patients. This was demonstrated by the fact that these patients had an enlarged spleen and an enlarged liver.
The polymerase chain reaction (PCR) done in real time was used to carry out the molecular detection. The goal is to produce the normal curve and to achieve exact measurement of the sample data, cDNA in a known quantity was incorporated into the analysis at the right volume. cDNA was manufactured using the RNA that was isolated from the sample in the laboratory. This allowed for the production of the standard curve as well.
Before beginning the process of cDNA synthesis, an RNA sample was collected from a Nanodrop for the purposes of analysis. For the aim of determining the nucleic acid content of the sample under analysis, both in terms of purity and amount, the technique of choice is one that is known as nanodrop RNA. This approach involves dropping small amounts of RNA into a solution. A method that is known as spectrophotometry can be utilised in order to determine the amount of light that is taken in by an organism. If RNA and DNA both have an absorbance in the 260 nm spectrum, then salt and protein should each have an absorbance in the 230 nm range and the 280 nm spectrum, respectively. We were able to assess whether or not the RNA that was extracted from salt was of a quality that was adequate by comparing the wavelengths at 260 and 230 nanometers. The ratio of 260 nm to 280 nm was measured for the protein in order to determine whether or not all of the RNA had been successfully extracted from the protein. This was done to determine whether or not all of the RNA had been successfully extracted. The ratio of 260 to 230 nanometers is greater than 1.8. As demonstrated in the figure, the sample in question is free of any impurities and does not include even the tiniest amounts of salt that might be regarded as a contaminant. In addition, the fact that the protein ratio is more than 1.8 gives rise to the hypothesis that the sample of RNA that we have access to does not contain an adequate amount of protein impurity.
A wide range of specialized instruments was utilized in order to be successful in achieving this objective. In order to detect a particular chromosomal mutation, a
The BCR-ABL experiment was carried out using specific BCR primers. This was done in order to locate the mutation. This made it possible to determine which mutation had occurred. It was discovered that the CT value was somewhere in the neighbourhood of 25 cycles in each of the fifteen positive samples, which served as verification that this was in fact the case. We also carried out a control experiment using water as a negative control so that we could remove the possibility of any errors arising during the process. Calculations were performed, that generated a standard curve. Following those calculations, the data were examined per the method portrayed in the figures. The samples were put through both an intra-assay and an inter-assay, and the findings of both types of testing were tallied up and analyzed. In order to achieve the most precise result possible for the CML and ALL mutations, the tests were carried out on the same sample multiple times at varying intervals of time. Samples were run with gaps of a few hours to several days in between them It was revealed that there was approximately less than a five per cent discrepancy in the results of each inspection when the same samples were analyzed at many different dates. In the end, we got the data, which showed that 15 Philadelphia p190 positive ALL tested positive while 45 of the CML samples, positive for the Philadelphia chromosome but negative for p190, tested negative along with 15 Philadelphia negative ALL samples using the same method. This demonstrates a total of one hundred per cent sensitivity and specificity.
Figure 1
Distribution of All and CMLL with Regards to Philadelphia Chromosome.
Figure 2
Age of the Participants Diagnosed with
Figure 3
Age of the Participants Diagnosed with CML.
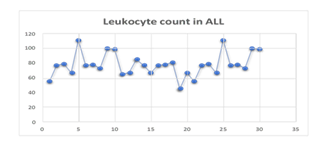
Figure 4
Leukocyte Count of the Participants Diagnosed with ALL.
Discussion
Acute lymphoblastic leukaemia (ALL), which has a copy of the Philadelphia chromosome, has been diagnosed in people of various ages. In two cases, children of various ages, beginning with those under the age of ten, were affected by the ailment (Guan 2021). Not only was it found most commonly in younger individuals, but it was also detected most frequently in persons who were at least forty years old.
The specimens were inspected at different time periods in order to get accurate results. Several of the tests on the specimens were done over a 2-3 day span, others were done in a lesser amount of time. (Saha 2023). The final results show that 15 ALL cases were determined to be negative using the same approach, along with 45 CML cases which were also negative for the Philadelphia chromosome using the same techniques. This displays a 100% total sensitivity (Mejía-Aranguré 2021).
A specific real-time PCR approach was used in this study to identify the specific Philadelphia mutation in ALL. Similar RT-PCR protocols were used by Emig, M. et al. While using the Light Cycler technology, Emig, M. et al., intended to develop a quick and accurate RT-PCR method for detecting and quantifying BCR-ABL fusion transcripts (Saygin 2022, Tierens 2021). This gadget uses fluorescence resonance energy transfer (FRET) to combine quick thermocycling with online measurement of PCR product generation. In a prospective study, 65 individuals with ALL were examined using the RT-PCR method and were given BCR-ABL transcript measurements before and after receiving an allogeneic transplant. They stated that the endogenous reference (GAPDH), was used to measure and normalise the expression of the BCR-ABL transcript (Stella 2019, Fukutsuka 2022). The real-time PCR approach was used by Beelen, D. W. et al., to track the effectiveness of several immunotherapies, in 10 ALL patients who had relapsed. The findings of the quantitative analysis of BCR-ABL transcripts were highly indicative of the clinical effect of the different applied immunotherapies (Chong 2021, Smith 2021). The innovative real-time PCR technique seems to be an effective tool for the early identification of recurrence after an allogeneic transplant in patients with the BCR-ABL transcript. Early treatment decisions are made possible by its capacity to differentiate between molecular and cytogenetic recurrence (P0.001) ( Piedimonte 2019).
SCT, contemporary PCR methods offer a fast, reliable, and highly sensitive tool, which may help identify individuals at high risk for leukemic recurrence (Chen 2022). Using a pool of samples from three confirmed ALL victims (cases 7, 20, and 22) and a diluent pool of specimens from healthy people, researchers assessed the analytical efficacy of the BCR-ABL1 p190 PCR test. A specimen collection from 3 ALL patients was used to assess the analytical efficacy of the BCR-ABL1 p190 PCR assay. The results of the serial dilutions demonstrated exceptional linearity, dependability, and accuracy of up to 0.001%. Since the negative samples lacked any kind of background, the detection of even a single droplet was considered very significant (Pfeifer 2019). In sixteen of the twenty-six patients who were subjected to follow-up monitoring, polymerase chain reaction (PCR) analysis was performed at a total of 117 time points. All twenty-six instances of ALL had their PCR testing done satisfactorily while they were unwell. At the first signs of the illness, there was a range in the number of BCR-ABL1 fusion transcripts, with a median proportion of 101–103 BCR-ABL1 copies. It is important to note that ALL patients with an extra copy of the Ph chromosome or overexpression of the BCR-ABL1 gene had high levels of the transcript, and researchers verified that the median value in these last instances was significantly distinct from what was discovered for the entire group (Pfeifer 2019).
Although TKI therapy for older patients with Ph+ ALL has significantly altered the trajectory of the illness, with 95% to 100% of the treated group experiencing complete responses, the vast majority of patients relapse rapidly, even after hematopoietic SCT (Podvin 2022). However, a standard method for measuring the BCR-ABL1 fusion transcript is still lacking. Lab-based qPCR is often used for MRD molecular monitoring in patients with Ph+ ALL. By using both calibrators and laboratory conversion factors, the standardisation of experimental and analytical processes for BCR-ABL1 p210 transcript observing in CML produced positive findings ( Zuna 2022). However, due to a significant amount of heterogeneity included in the qPCR technique, the effectiveness of BCR-ABL1 transcript quantification may fluctuate significantly between
laboratories.
De Haas et al. and Fronkova et al. compared the MRD outcomes of RT-qPCR of fusion genes and qPCR of Ig/TCR rearrangements in 13 and 28 cases, respectively. The two qPCR-based approaches were shown to have a positive association in ALL, but a negative correlation in BCR-ABL1 ALL. Using genomic DNA as a target, the same association was seen between fusion transcripts-MRD and Ig/TCR-MRD experiments (Spearman correlation, 0.899 for ETV6-RUNX1 ALL and 0.62 for BCR-AB 1 ALL). ETV6-RUNX1 ALL (87.1%) had a greater number for concordance than BCR-ABL1 ALL (70.6%) in terms of showing the correlation of outcomes between the two qPCR-based techniques. (Shires 2020).
Consistently high levels of agreement (93.3%, value of 0.842) between 6 colour flow cell measurements for immunophenotypes and RT-qPCR for fusion genes, associated with leukaemia regardless of the choice of assaying technique. In contrast to allele-specific oligonucleotide-qPCR for clonal Ig/TCR in individual patients, a single primer set may be used for RT-qPCR of fusion transcripts in all individuals carrying TCF3-PBX1. More time and money may be saved by using this method. Six-colour flow cytometry is only as sensitive as RT-qPCR for TCF3-PBX1, which may reach a sensitivity of 105. TCF3-PBX1 ALL MRD monitoring is the most cost-effective when performed via RT-qPCR for fusion transcripts ( Correia 2021).
When viewed as a whole, our findings showed that each fusion transcript had a tendency toward using a certain VH. Despite the fact that the two qPCR-based in TCF3-PBX1 ALL, approaches showed excellent MRD concordance, RT-qPCR was shown to be the most cost- and time-effective method, whilst Ig/TCR-qPCR was found to be the most reliable method for guiding MRD-adapted therapy in BCR-ABL1 ALL (Shor 2019).
Traditional clinical laboratories can measure the BCR-ABL1 p190 fusion transcript using PCR without the need for calibrators, standard curves, or laboratory conversion factors at the time of ALL identification and during MRD assessment. Additional testing of the p190 PCR assay in a wider cohort of patients will help clarify its clinical prognostic value in Ph+ ALL MRD monitoring and establish standardized criteria for data interpretation (Della 2023).
ALL, which has a copy of the Philadelphia chromosome, has been diagnosed in people of various ages. The majority of studies have found a correlation between ageing and worse clinical results and a higher incidence of Ph-like ALL. Although one small trial found no difference in outcomes between Ph-like and non-Ph-like ALL patients, 15 per cent of Ph-like ALL patients got HSCT in first remission owing to poor MRD response. Although most cases of B-ALL are diagnosed in children and young adults, new research shows that the Ph-like subtype is very frequent in individuals aged 40 and over. In adults, fusions are more common than they are in children, and the incidence of other sentinel genetic changes varies widely between the two age groups. Other risk factors for treatment failure and recurrence in Ph-like ALL include a greater white blood cell count, a higher incidence of minimum residual illness after induction, and older age. Patients who resemble a Ph yet have a high end-of-induction Those with MRD have an exceptionally terrible prognosis, but patients with Ph-like symptoms are not likely to survive regardless of genetic alteration. It is well established that certain patients with Ph-like ALL exhibit blatant induction failure (Bartram 2020).
In order to detect ABL-class fusions in ph-ALL patients, a battery of multiplex RT-PCR tests and DNA sequencing is being developed. Many studies have shown multiple underlying genetic abnormalities in Ph-like ALL, many of which may be addressed by already available small molecule inhibitors, and many of these abnormalities have been related to poor survival in both children and adults. Clinical activity of these drugs has been documented in anecdotal accounts, and currently, single-arm studies are being conducted to see whether adding targeted medicines like dasatinib and ruxolitinib to regular chemotherapy helps.
Conclusion
Increased identification of BCR-ABL1 was demonstrated using real-time polymerase chain reaction. Real-time qPCR is an effective technique for disease tracking in patients with acute lymphoblastic leukaemia due to better BCR-ABL1 p190 detection and the possibility for improved uniformity across numerous labs.
References
- Huang, Y. J., Kuo, M. C., Jaing, T. H., Liu, H. C., Yeh, T. C., & Chen, S. H. (2021). Comparison of two quantitative PCR–based assays for detection of minimal residual disease in B-precursor acute lymphoblastic leukaemia harbouring three major fusion transcripts. The Journal of Molecular Diagnostics. 23(10), 1373-9
- Martinez, R. J., Kang, Q., Nennig, D., Bailey, N. G., Brown, N. A., & Betz, B. L. (2022). One- Step Multiplexed Droplet Digital Polymerase Chain Reaction for Quantification of p190 BCR-ABL1 Fusion Transcript in B-Lymphoblastic Leukemia. Archives of Pathology & Laboratory Medicine. 146(1), 92-100.
- Beldinanzi, M., Della, S. I., Cardinali, D., Bellomarino, V., Elia, L., & Matarazzo, M. (2022). Digital Droplet PCR for Minimal Residual Disease Assessment in Philadelphia-Positive Acute Lymphoblastic Leukemia Using IG/TR Genes and BCR/ABL1 as Markers. Preliminary Results of a Comparative Analysis . Blood. 140, 11866-7.
- Ansuinelli, M., Della, S. I., Lauretti, A., Elia, L., Siravo, V., & Messina, M. (2021). Applicability of droplet digital polymerase chain reaction for minimal residual disease monitoring in Philadelphia-positive acute lymphoblastic leukaemia. Hematological Oncology. 39(5), 680-6.
- Guan, Y., Zhang, M., Zhang, W., Wang, J., Shen, K., & Zhang, K. (2021). Clinical Utility of Droplet Digital PCR to Monitor BCR-ABL1 Transcripts of Patients With Philadelphia Chromosome–Positive Acute Lymphoblastic Leukemia Post-chimeric Antigen Receptor19/22 T-Cell Cocktail Therapy. Frontiers in Oncology. 11, 646499.
- Saha, J., Gopinath, V., Nair, C. K., & Roshan, D. (2023). Identification of rare atypical BCR- ABL1 transcript: A case report.
- Saygin, C., Cannova, J., Stock, W., & Muffly, L. (2022). Measurable residual disease in acute lymphoblastic leukaemia: methods and clinical context in adult patients. Haematologica. 107(12), 2783-93.
- Tierens, A., Stockley, T. L., Campbell, C., Fulcher, J., Leber, B., & McCready, E. (2021). Consensus recommendations for MRD testing in adult B-cell acute lymphoblastic leukaemia in Ontario. Current Oncology. 28(2), 1376-87.
- Stella, S., Gottardi, E. M., Favout, V., Barragan, G. E., Errichiello, S., & Vitale, S. R. (2019). The q-lamp method represents a valid and rapid alternative for the detection of the bcr-abl1 rearrangement in Philadelphia-positive leukaemias . International Journal of Molecular Sciences. 20(24), 6106.
- Fukutsuka, K., Kuramura, A., Nakagawa, M., Takahashi, R., Chagi, Y., & Nakagawa, M. (2022). Philadelphia chromosome-positive acute lymphoblastic leukaemia carrying the p230 μ-BCR-ABL1 fusion gene. Tenri Medical Bulletin, 25(1), 29-40.
- Chong, S. L., Asnawi, A. W. A., Leong, T. S., Tan, J. T., Law, K. B., & Hon, S.L. (2021). Impact of timely BCR-ABL1 monitoring before allogeneic stem cell transplantation among patients with BCR-ABL1-positive B-acute lymphoblastic leukaemia. Blood research. 56(3), 175-83.
- Smith, B. M., Brewer, D., Druker, B. J., & Braun, T. P. (2021). Navigating challenges in monitoring chronic myeloid leukaemia with multiple BCR-ABL1 transcripts. Case Reports in Oncology. 14, 1707-11.
- Piedimonte, M., Ottone, T., Alfonso, V., Ferrari, A., Conte, E., & Divona, M. (2019). A rare BCR-ABL1 transcript in Philadelphia- positive acute myeloid leukaemia: case report and literature review. BMC cancer. 19, 1-6.
- Chen, Y., Lu, Q. Y., Lu, J. Y., & Hong, X. L. (2022). Primary isolated central nervous system acute lymphoblastic leukaemia with BCR-ABL1 rearrangement: A case report. World Journal of Clinical Cases. 10(13), 4242.
- Pfeifer, H., Cazzaniga, G., Van der Velden, V., Cayuela, J., Schäfer, B., & Spinelli, O. (2019). Standardisation and consensus guidelines for minimal residual disease assessment in Philadelphia-positive acute lymphoblastic leukaemia (Ph+ ALL) by real-time quantitative reverse transcriptase PCR of e1a2 BCR-ABL1. Leukaemia. 33(8), 1910-22.
- Podvin, B., Guermouche, H., Roynard, P., Goursaud, L., Berthon, C., & Ouafi, M. (2022). Subclonal acquisition of a BCR:: ABL1 fusion in chronic myelomonocytic leukaemia. Annals of Hematology, 101(9), 2093-5.
- Zuna, J., Hovorkova, L., Krotka, J., Koehrmann, A., Bardini, M., & Winkowska, L. (2022). Minimal residual disease in BCR:: ABL1- positive acute lymphoblastic leukaemia: different significance in typical ALL and in CML-like disease. Leukaemia, 1-9.
- Shires, K., & Rust, A. (2020). Novel multiplex RT-PCR assay to detect BCR/ABL mRNA variants. The Journal of Medical Laboratory Science and Technology of South Africa. 2(1), 29-35.
- Correia, R. P., Bento, L. C., Sousa, F. A., Barroso, R. S., Campregher, P. V., & Bacal, N. S. (2021). How I investigate minimal residual disease in acute lymphoblastic leukaemia. International Journal of Laboratory Hematology. 43(3), 354-63.
- Short, N. J., Jabbour, E., Albitar, M., de Lima, M., Gore, L., & Jorgensen, J. (2019). Recommendations for the assessment and management of measurable residual disease in adults with acute lymphoblastic leukaemia: a consensus of North American experts. American journal of haematology, 94(2), 257- 65.
- Della Starza, I., De Novi, L. A., Elia, L, Bellomarino, V., Beldinanzi, M., & Soscia, R, (2023). Optimizing Molecular Minimal Residual Disease Analysis in Adult Acute Lymphoblastic Leukemia. Cancers. 15(2), 374.
Cite this article
-
APA : Munir, G., Gulrukh., & Tariq, H. (2022). Development of Real-Time Quantitative PCR Assay for Detection and Monitoring of BCR ABL1 P190 Transcript in Acute Lymphoblastic Leukemia. Global Drug Design & Development Review, VII(IV), 39-47. https://doi.org/10.31703/gdddr.2022(VII-IV).05
-
CHICAGO : Munir, Gulshan, Gulrukh, and Hafsa Tariq. 2022. "Development of Real-Time Quantitative PCR Assay for Detection and Monitoring of BCR ABL1 P190 Transcript in Acute Lymphoblastic Leukemia." Global Drug Design & Development Review, VII (IV): 39-47 doi: 10.31703/gdddr.2022(VII-IV).05
-
HARVARD : MUNIR, G., GULRUKH. & TARIQ, H. 2022. Development of Real-Time Quantitative PCR Assay for Detection and Monitoring of BCR ABL1 P190 Transcript in Acute Lymphoblastic Leukemia. Global Drug Design & Development Review, VII, 39-47.
-
MHRA : Munir, Gulshan, Gulrukh, and Hafsa Tariq. 2022. "Development of Real-Time Quantitative PCR Assay for Detection and Monitoring of BCR ABL1 P190 Transcript in Acute Lymphoblastic Leukemia." Global Drug Design & Development Review, VII: 39-47
-
MLA : Munir, Gulshan, Gulrukh, and Hafsa Tariq. "Development of Real-Time Quantitative PCR Assay for Detection and Monitoring of BCR ABL1 P190 Transcript in Acute Lymphoblastic Leukemia." Global Drug Design & Development Review, VII.IV (2022): 39-47 Print.
-
OXFORD : Munir, Gulshan, Gulrukh, , and Tariq, Hafsa (2022), "Development of Real-Time Quantitative PCR Assay for Detection and Monitoring of BCR ABL1 P190 Transcript in Acute Lymphoblastic Leukemia", Global Drug Design & Development Review, VII (IV), 39-47
-
TURABIAN : Munir, Gulshan, Gulrukh, and Hafsa Tariq. "Development of Real-Time Quantitative PCR Assay for Detection and Monitoring of BCR ABL1 P190 Transcript in Acute Lymphoblastic Leukemia." Global Drug Design & Development Review VII, no. IV (2022): 39-47. https://doi.org/10.31703/gdddr.2022(VII-IV).05